Correlation Engine Highlighting Service  Highlight your site
Highlight your site
Contents
- Overview of Highlighting Your Own Pages
- How the Correlation Engine Highlighting Service Works
- Information Transmitted to Correlation Engine
- Instructions for Adding The Correlation Engine Highlighting Service to your Page
- Example
Overview of Highlighting Your Own Pages
The instructions on this page will enable the Enterprise edition of the Correlation Engine Highlighting Service. For a list of differences from the public Correlation Engine Highlighting Services, see the Enterprise section of the Correlation Engine Highlighting Service FAQ.
By adding just one line of JavaScript to your web page, you can augment it with the Correlation Engine Highlighting Service. Any user visiting your page will see biological terms underlined. When the user moves the mouse over a term, more information about that term is displayed, including related literature, other related terms, and additional information about that term. The following paragraph shows what this highlighting looks like. (It may take a few seconds after the page loads before the highlighting is visible). Note that this example uses the public Correlation Engine Highlighting Service, not the Enterprise edition.
ERBB2: This gene encodes a member of the epidermal growth factor (EGF) receptor family of receptor tyrosine kinases. This protein has no ligand binding domain of its own and therefore cannot bind growth factors. However, it does bind tightly to other ligand-bound EGF receptor family members to form a heterodimer, stabilizing ligand binding and enhancing kinase-mediated activation of downstream signalling pathways, such as those involving mitogen-activated protein kinase and phosphatidylinositol-3 kinase. Allelic variations at amino acid positions 654 and 655 of isoform a (positions 624 and 625 of isoform b) have been reported, with the most common allele, Ile654/Ile655, shown here. Amplification and/or overexpression of this gene has been reported in numerous cancers, including breast and ovarian tumors. Alternative splicing results in several additional transcript variants, some encoding different isoforms and others that have not been fully characterized. [provided by RefSeq]
Move your mouse over any of the underlined terms in the above paragraph for more information about that term. You can also see a larger example based on a Wikipedia page.
The Correlation Engine Highlighting API can also be used to create a bookmark that can be used to highlight any page on the web. You can learn more about it here.
How the Correlation Engine Highlighting Service Works
After a page is loaded, the Correlation Engine highlighting Service is invoked. Normally, this happens in the onload event as part of the JavaScript onload event, but if you need to, you can defer the highlighting by adding a few additional lines of JavaScript (for example, if you want to wait until after an AJAX update).
The script reads the content of the page, and sends it to Correlation Engine for processing. The Correlation Engine servers will parse the text, looking for terms of biological significance. Those terms will be sent back to your browser, where the JavaScript will look for those terms and highlight them.
When you move your mouse over a term, that term is sent to the Correlation Engine Server. That term is correlated with the text of the page, to find publications, clinical trials, Genomic studies, and other information related to that term, in the context of the original page.
Information Transmitted to Correlation Engine
In order to locate biological terms in your page, and to determine the related information, the Correlation Engine Highlighting Service sends the contents of the entire page to Correlation Engine. Correlation Engine uses this information only to provide The Correlation Engine Highlighting Service. Correlation Engine's handling, possession, and use of this information complies with the Correlation Engine terms of service and privacy policy. Because the contents of the entire page are transmitted to Correlation Engine, you should refrain from using this service when viewing pages which contain sensitive information, including personal information.
Instructions for Adding The Correlation Engine Highlighting Service to your Page
Adding the Correlation Engine Highlighting Service requires the following steps:
- Obtain a Correlation Engine Highlighting Service key and agree to the terms of service. Obtain your key here.
- Add a line of JavaScript to each pages. This line will be provided to you once you obtain your key.
- Test your pages.
Step One: Obtain a Correlation Engine Highlighting Service Key
The Correlation Engine Highlighting Service requires a Correlation Engine Highlighting Service Key.
Get your Correlation Engine Highlighting Service Key
After you have your key, you will return to this page, where you can continue.
Step Two: Add the JavaScript near the end of your web page.
Add one line of JavaScript to the <body> of your web page. This line will be provided to you once you obtain your Correlation Engine Highlighting Service Key. It must be in the <body> section; if it is in the <head>, highlighting may not take place. We recommend adding it near the bottom of the <body>, e.g. just before the </body>, to avoid slowing down the display of your page. If you use a common include or template to display page footers, you should add it there. If your page uses framesets, you should add the line to the frames that you wish to highlight, rather than adding it to the frameset.
Step Three: Test your page.
Load your modified page into a browser. You should see the highlighting on your page. If you don't, please see the highlighting FAQ
Example
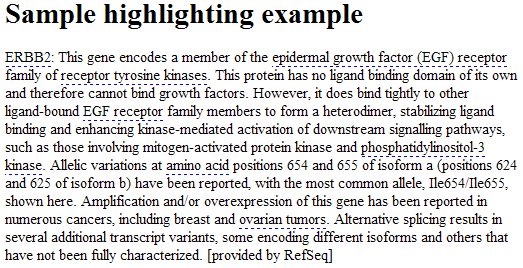
Here is a sample web page with the embedded JavaScript:
<html>
<head>
<title>Correlation Engine Highlighting Example</title>
</head>
<body>
<h1>Sample highlighting example</h1>
ERBB2: This gene encodes a member of the epidermal growth factor (EGF) receptor family of receptor tyrosine kinases.
This protein has no ligand binding domain of its own and therefore cannot bind growth factors. However,
it does bind tightly to other ligand-bound EGF receptor family members to form a heterodimer, stabilizing
ligand binding and enhancing kinase-mediated activation of downstream signalling pathways, such as those
involving mitogen-activated protein kinase and phosphatidylinositol-3 kinase. Allelic variations at amino
acid positions 654 and 655 of isoform a (positions 624 and 625 of isoform b) have been reported, with the
most common allele, Ile654/Ile655, shown here. Amplification and/or overexpression of this gene has been
reported in numerous cancers, including breast and ovarian tumors. Alternative splicing results in several
additional transcript variants, some encoding different isoforms and others that have not been fully
characterized. [provided by RefSeq]
<!--Include the script at the end of the body-->
<script type='text/javascript' src='http://api.nextbio.com/b/nbapi/include.nbs?type=js&name=genhighlight&
</body>
</html>
The resulting page looks like this: